Research
We aim to elucidate the functional expression mechanisms of proteins and functional RNAs related to diseases (such as AIDS, cancer, and prion diseases) and the pluripotency of neural cells based on their three-dimensional structures.
Once the mechanisms are elucidated, it becomes possible to modify these molecules into more functional and useful forms based on the obtained knowledge, providing valuable insights for drug development. In parallel with the structural approach, we also verify the functions of these molecules in vivo using cultured cells and other biological systems.
We are also conducting structural and dynamic analyses of non-edible lignocellulosic biomass. Based on our findings, we are developing new methods to extract valuable substances from lignocellulosic biomass that can serve as raw materials for bioenergy (such as bioethanol) and various products (such as bioplastics and cosmetics). Ultimately, our research aims to contribute to a paradigm shift toward an environmentally sustainable biorefinery.
The preparation of samples for research and the modification of molecules to enhance their utility are carried out using advanced genetic engineering techniques. Additionally, the determination of three-dimensional molecular structures is conducted using three nuclear magnetic resonance (NMR) spectrometers.
Verification using cultured cells is conducted by combining molecular biological techniques such as RNA interference (RNAi) with fluorescence observation. The specific research topics are outlined below.
01. A Structural Biology Approach to Enhancing Enzymatic Conversion of Lignocellulosic Biomass

The utilization of lignocellulosic biomass, which does not compete with food resources, holds great potential for reducing dependence on fossil fuels.
Our research focuses on the structural biology of cellulolytic and ligninolytic enzymes derived from wood-decaying fungi, which play a key role in breaking down lignocellulosic components. Additionally, we are developing solution-state NMR techniques for analyzing lignocellulosic biomass, enabling us to track each stage of chemical structural changes and degradation processes in detail—providing insights that are difficult to obtain with other analytical methods.
By integrating enzyme science with advanced NMR techniques, we aim to establish novel utilization strategies for lignocellulosic biomass, enabling the efficient production of bioenergy and valuable chemicals. Ultimately, our goal is to drive a paradigm shift toward a next-generation biorefinery independent of fossil fuels.
02. RNA molecules (RNA aptamers) that capture prion proteins with high affinity
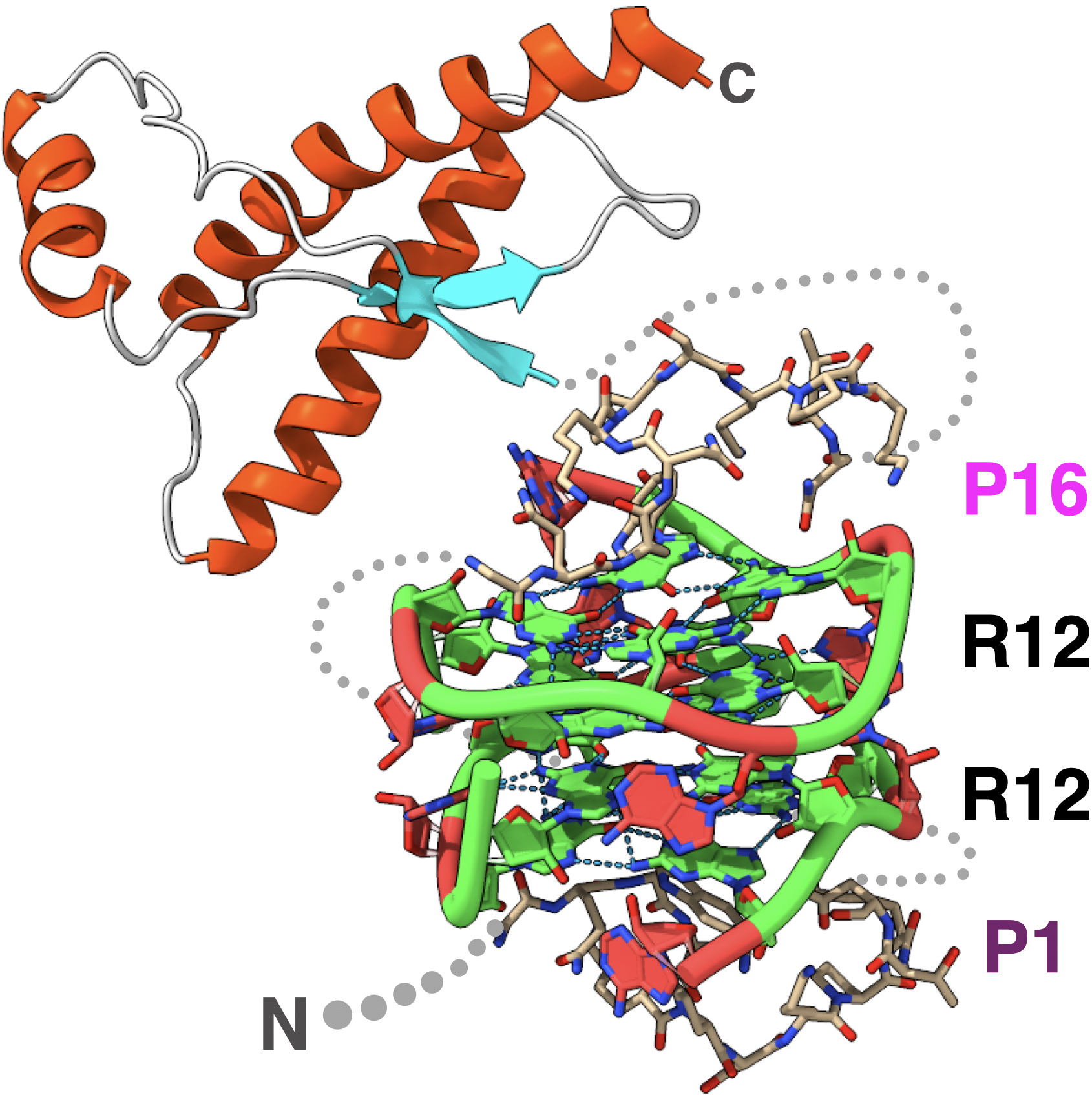
The three-dimensional structure of this RNA aptamer was determined using NMR, and it was found to form a unique four-stranded structure consisting of four guanine bases and two adenine bases, creating a hexad structure.
Additionally, structural analysis of the RNA aptamer-prion protein complex revealed the mechanism by which the high capture ability is exhibited.
Furthermore, experiments using living cells demonstrated that this RNA aptamer has anti-prion disease activity.
03. The human protein APOBEC3G with anti-HIV activity
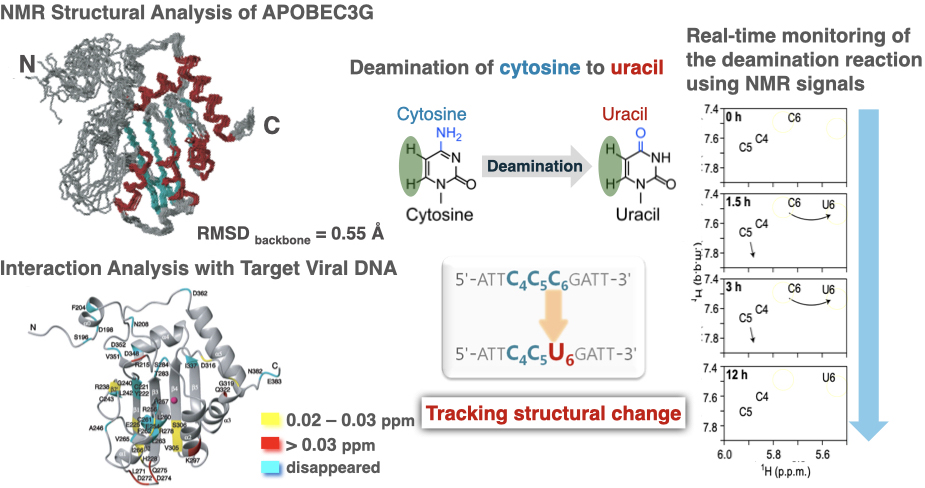
APOBEC3G acts on the negative-strand DNA of HIV, deaminating cytosine to convert it into uracil, thereby disrupting the genomic information of HIV and exhibiting anti-HIV activity. We determined the three-dimensional structure of this protein and its interaction with the target DNA using NMR spectroscopy.
Furthermore, we were the first in the world to successfully monitor the deamination reaction in real-time using NMR signals. The insights gained from this study are expected to be useful for the development of anti-HIV drugs.
04. Structural Biology Studies of Nucleic Acids in Living Cells Using In-Cell NMR Spectroscopy
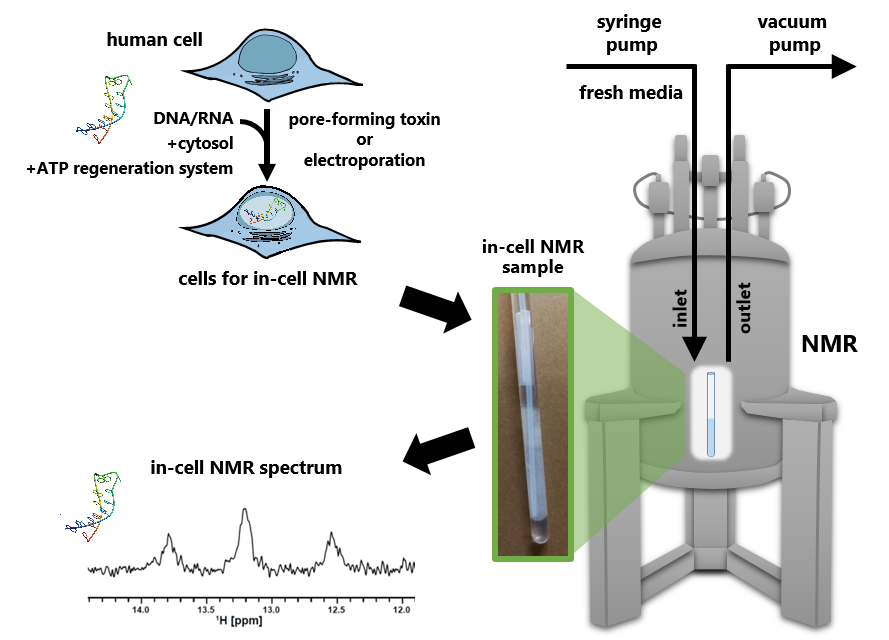
The intracellular environment of living cells is characterized by a highly dense and heterogeneous molecular crowding condition. Under such conditions, the structures, dynamics, and intermolecular interactions of proteins and nucleic acids are different from those observed in dilute solutions under in vitro conditions. However, methodologies that enable us to investigate these phenomena remain limited.
To address this, we have been developing in-cell NMR spectroscopy, a technique that enables the direct acquisition of NMR spectra of proteins and nucleic acids within living cells. Using this approach, we have demonstrated the formation of unique nucleic acid structures, such as a triplex and a G-quadruplex, as well as the formation of complexes between an RNA aptamer and its target molecule in living human cells.
Furthermore, our studies have revealed that in human living cells, highly stable structures, G-quadruplex, undergo more frequent dissociation compared to in vitro conditions. These findings provide new insights into the regulatory mechanisms of nucleic acid structures within the intracellular environment.
05. The protein Musashi, which is involved in maintaining the undifferentiated state (pluripotent state) of neural progenitor cells.
Musashi maintains the undifferentiated state by binding to the 3 untranslated region (UTR) of target gene mRNAs and inhibiting their translation.
Using advanced NMR techniques, including paramagnetic relaxation enhancement and residual dipolar couplings, we successfully extracted long-range (~30 Å) structural information and determined the three-dimensional structure of the Musashi-target RNA complex.
Additionally, we are conducting analyses of its interactions with poly(A)-binding proteins. These studies are progressively revealing the complete mechanism by which Musashi suppresses the translation of specific genes.
06. Interaction between non-coding RNA and proteins
We are investigating the functional mechanisms of non-coding RNAs involved in transcriptional repression and telomere length regulation, along with their interacting proteins, using a structural life science approach.
